This topic is under construction for the 24.3 (March 2024) release of LabKey Server. For current documentation of this feature, click here.
The R language is a good choice for writing assay transformation scripts, because it contains a lot of built-in functionality for manipulating tabular data sets.
General information about creating and using transformation scripts can be found in this topic:
Transform Scripts. This topic contains information and examples to help you use R as the transformation scripting language.
Include Other Scripts
If your transform script calls other script files to do its work, the normal way to pull in the source code is using the source statement, for example:
source("C:\lktrunk\build\deploy\files\MyAssayFolderName\@files\Utils.R")To keep dependent scripts so that they are easily moved to other servers, it is better to keep the script files together in the same directory. Use the built-in substitution token "${srcDirectory}" which the server automatically fills in to be the directory where the called script file (the one identified in the
Transform Scripts field) is located, for example:
source("${srcDirectory}/Utils.R");Connect Back to the Server from an R Transform Script
Sometimes a transform script needs to connect back to the server to do its job. One example is translating lookup display values into key values. The Rlabkey library available on CRAN has the functions needed to connect to, query, and insert or update data in the local LabKey Server where it is running. To give the connection the right security context (that of the current user), the assay framework provides the substitution token ${rLabkeySessionId}. Including this token on a line by itself near the beginning of the transform script eliminates the need to use a config file to hold a username and password for this loopback connection. It will be replaced with two lines that looks like:
labkey.sessionCookieName = "JSESSIONID"
labkey.sessionCookieContents = "TOMCAT_SESSION_ID"
where TOMCAT_SESSION_ID is the actual ID of the user's HTTP session.
Debug an R Transform Script
You can load an R transform script into the R console/debugger and run the script with debug(<functionname>) commands active. Since the substitution tokens described above ( ${srcDirectory} , ${runInfo}, and ${rLabkeySessionId} ) are necessary to the correct operation of the script, the framework conveniently writes out a version of the script with these substitutions made, into the same subdirectory where the runProperties.tsv file is found. Load this modified version of the script into the R console.
Example Script
Scenario
Suppose you have the following Assay data in a TSV format:
| SpecimenID | Date | Score | Message |
|---|
| S-1 | 2018-11-02 | 0.1 | |
| S-2 | 2018-11-02 | 0.2 | |
| S-3 | 2018-11-02 | 0.3 | |
| S-4 | 2018-11-02 | -1 | |
| S-5 | 2018-11-02 | 99 | |
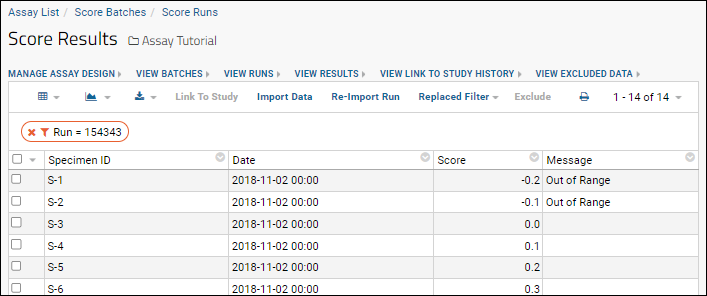
You want a transform script that can flag values greater than 1 and less than 0 as "Out of Range", so that the data enters the database in the form:
| SpecimenID | Date | Score | Message |
|---|
| S-1 | 2018-11-02 | 0.1 | |
| S-2 | 2018-11-02 | 0.2 | |
| S-3 | 2018-11-02 | 0.3 | |
| S-4 | 2018-11-02 | -1 | Out of Range |
| S-5 | 2018-11-02 | 99 | Out of Range |
exampleTransform.R
The following R transform script accomplishes this and will write to the Message column if it sees out of range values:
library(Rlabkey)
###################################################################################
# Read in the run properties. Important run.props will be set as variables below #
###################################################################################
run.props = labkey.transform.readRunPropertiesFile("${runInfo}");
###########################################################################
# Choose to use either the raw data file (runDataUploadedFile) #
# or the LabKey-processed TSV file (runDataFile) if it can be created. #
# Uncomment one of the two options below to set the run.data.file to use. #
###########################################################################
# Use the original file uploaded by the user. (Use this if the assay framework fails to convert it to an TSV format.)
#run.data.file = labkey.transform.getRunPropertyValue(run.props, "runDataUploadedFile");
# Use the file produced after the assay framework converts the user uploaded file to TSV format.
run.data.file = labkey.transform.getRunPropertyValue(run.props, "runDataFile");
##########################################################
# Set the output and error files as separate variables. #
##########################################################
run.output.file = run.props$val3[run.props$name == "runDataFile"];
error.file = labkey.transform.getRunPropertyValue(run.props, "errorsFile");
####################################################################################
# Read in the results content to run.data - this example supports several formats. #
####################################################################################
if (grepl("\\.tsv$", run.data.file)) {
run.data = read.delim(run.data.file, header=TRUE, sep="\t", stringsAsFactors = FALSE)
} else if (grepl("\\.csv$", run.data.file)) {
run.data = read.csv(run.data.file, header=TRUE, stringsAsFactors = FALSE)
} else if (grepl("\\.xlsx$", run.data.file)) {
run.data = read_excel(run.data.file, sheet = 1, col_names = TRUE)
} else {
stop("Unsupported file type. Please provide a TSV, CSV, or Excel file.")
}
# If you know you only need the TSV format, you could simplify the above to use only:
#run.data = read.delim(run.data.file, header=TRUE, sep="\t", stringsAsFactors = FALSE);
###########################################################
# Transform the data. Your transformation code goes here. #
###########################################################
# If any Score value is less than 0 or greater than 1,
# then place "Out of Range" in the Message vector.
for(i in 1:nrow(run.data))
{
if (run.data$Score[i] < 0 | run.data$Score[i] > 1) {run.data$Message[i] <- "Out of Range"}
}
###########################################################
# Write the transformed data to the output file location. #
###########################################################
# write the new set of run data out to an output file
write.table(run.data, file=run.output.file, sep="\t", na="", row.names=FALSE, quote=FALSE);
writeLines(paste("\nProcessing end time:",Sys.time(),sep=" "));;
Setup
Before installing this example, ensure that an
R engine is configured on your server.
- Create a new folder of type Assay.
- Create an Assay Design (in the current folder) named "Score" with the following data fields. You can either enter them yourself or download and import this assay design: Score.xar
| Name | Data Type |
|---|
| SpecimenId | Text |
| Date | DateTime |
| Score | Decimal (floating point) |
| Message | Text |
- Download this R script: exampleTransform.R
- Edit the assay design to add the transform script.
- Select > Manage Assays and click Score.
- Select Manage Assay Design > Edit Assay Design.
- Click Add Script and select or drag and drop to add the script you downloaded. This will both upload it to the "@scripts" subdirectory of the file root, and add the absolute path to the assay design.
- Click Save.
- Import data to the Assay Design. Include values less than 0 or greater than 1 to trigger "Out of Range" values in the Message field. You can use this example data file: R Script Assay Data.tsv
- View the transformed results imported to the database to confirm that the R script is working correctly.
Related Topics