Examples 4, 5 & 6: Describe LCMS Experiments |
2024-04-18 |
This topic explores how a xar file can describe an MS2 analysis. Examples 4, 5, and 6 in the provided archive demonstrate these features.
Import XAR Files Using the Data Pipeline
If a xar.xml referencing MS2 data is imported via the Process and Import Data option on the data pipeline, and the file references are correct, the pipeline will automatically initiate an upload of the MS2 data. Note that this option is not available when importing via the file browser.
The xar.xml experiment description document is not intended to contain all of the raw data and intermediate results produced by an experiment run. Experimental data are more appropriately stored and transferred in structured documents that are optimized for the specific data and (ideally) standardized across machines and software applications. For example, MS2 spectra results are commonly transferred in "mzXML" format. In these cases the xar.xml file would contain a relative file path to the mzXML file in the same directory or one of its subdirectories. To transfer an experiment with all of its supporting data, the plan is that the folder containing xar.xml and all of its subfolder contents would be zipped up into an Experment Archive file with a file extension of "xar". In this case the xar.xml file acts like a "manifest" of the archive contents, in addition to its role as an experiment description document.
Connected Experiment Runs
Examples 4 and 5 are more “real world” examples. They describe an MS2 analysis that will be loaded into LabKey Server. These examples use the file Example4.mzXML in the XarTutorial directory. This file is the output of an LCMS2 run, a run which started with a physical sample and involved some sample preparation steps. The mzXML file is also the starting input to a peptide search process using X!Tandem. The search process is initiated by the Data Pipeline, and produces a file named Example4.pep.xml. When loaded into the database, the pep xml becomes an MS2 Run with its associated pages for displaying and filtering the list of peptides and proteins found in the sample. It is sometimes useful to think of the steps leading up to the mzXML file as a separate experiment run from the peptide search analysis of that run, especially if multiple searches are run on the same mzXML file. The Data Pipeline follows this approach.
To load both experiment runs, follow these steps.
- Download the file Example4.zip. Extract the files into a directory that is accessible to your LabKey Server, such as \\server1\piperoot\Example4Files. This folder will now contain a sample mzXML file from an LCMS2 run, as well as a sample xar.xml file and a FASTA file to search against.
- Because Example4 relies on its associated files, it must be loaded using the data pipeline (rather than the "upload xar.xml" button. Make sure the Data Pipeline is set to a root path above or including the Example4 folder.
- On the Pipeline tab, click Process and Upload Data.
- Check the box next to Example4.xar.xml and click Import Data. This loads a description of the experimental steps that produced the Example4.mzXML file.
- Return to the Process and Upload Data button on the Pipeline tab. This time select the Search for Peptides button next to the Example4.mzXML file. (Because these is already a xar.xml file with the same base name in the directory, the pipeline skips the page that asks the user to describe the protocol that produced the mzXML file.)
- The pipeline presents a dialog entitled Search MS2 Data. Choose the “Default” protocol that should appear in the dropdown. Press Search.
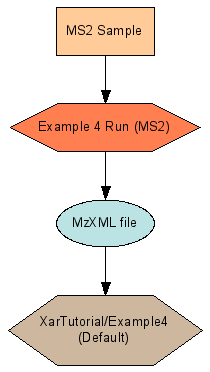
The peptide search process may take a minute or so. When completed, there should be a new experiment named “Default experiment for folder”. Clicking on the experiment name should show two runs belonging to it. When graphed, these two runs look like the following
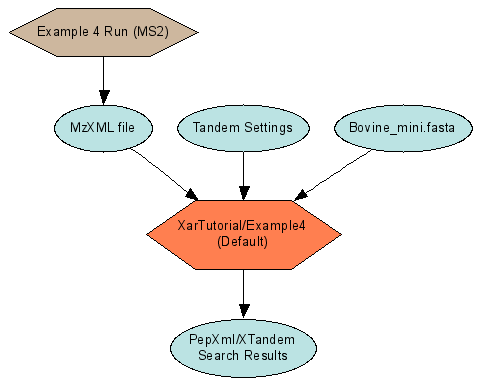
Connected runs for an MS2 analysis (Example 4)
|
|
|
|
Example 4 Run (MS2) Summary View |
XarTutorial/Example4 (Default) Summary View |
Referencing files for Data objects
The connection between the two runs is the Example4.mzXML file. It is the output of the run described by Example4.xar.xml. It is the input to a search run which has a xar.xml generated by the data pipeline, named XarTutorial\xtandem\Default\Example4.search.xar.xml. LabKey Server knows these two experiment runs are linked because the marked output of the first run is identified as a starting input to the second run. The file Example4.mzXML is represented in the xar object model as a Data object with a DataFileUrl property containing the path to the file. Since both of the runs are referring to the same physical file, there should be only one Data object created. The ${AutoFileLSID} substitution template serves this purpose. ${AutoFileLSID} must be used in conjunction with a DataFileUrl value that gives a path to a file relative to the xar.xml file’s directory. At load time the LabKey Server loader checks to see if an existing Data object points to that same file. If one exists, that object’s LSID is substituted for the template. If none exists, the loader creates a new Data object with a unique LSID. Sharing the same LSID between the two runs allows LabKey Server to show the linkage between the two, as in Figure 4.
Table 4: Example 4 LCMS Experiment description
|
Example4.xar.xml
The OutputDataLSID of the step that produces the mzXML file uses the ${AutoFileLSID} template. A second parameter, OutputDataFileTemplate, gives the relative path to the file from the xar.xml’s directory (in this case the file is in the same directory). |
<exp:Protocol rdf:about="${FolderLSIDBase}:ConvertToMzXML"> <exp:Name>Convert to mzXML</exp:Name> <exp:ApplicationType>ProtocolApplication</exp:ApplicationType> <exp:MaxInputMaterialPerInstance>0</exp:MaxInputMaterialPerInstance> <exp:MaxInputDataPerInstance>1</exp:MaxInputDataPerInstance> <exp:OutputMaterialPerInstance>0</exp:OutputMaterialPerInstance> <exp:OutputDataPerInstance>1</exp:OutputDataPerInstance> <exp:OutputDataType>Data</exp:OutputDataType> <exp:ParameterDeclarations> <exp:SimpleVal Name="ApplicationLSIDTemplate" OntologyEntryURI="terms.fhcrc.org#XarTemplate.ApplicationLSID" ValueType="String">${RunLSIDBase}:${InputLSID.objectid}.DoConvertToMzXML</exp:SimpleVal> <exp:SimpleVal Name="ApplicationNameTemplate" OntologyEntryURI="terms.fhcrc.org#XarTemplate.ApplicationName" ValueType="String">Do conversion to MzXML</exp:SimpleVal> <exp:SimpleVal Name="OutputDataLSIDTemplate" OntologyEntryURI="terms.fhcrc.org#XarTemplate.OutputDataLSID" ValueType="String">${AutoFileLSID}</exp:SimpleVal> <exp:SimpleVal Name="OutputDataFileTemplate" OntologyEntryURI="terms.fhcrc.org#XarTemplate.OutputDataFile" ValueType="String">Example4.mzXML</exp:SimpleVal> <exp:SimpleVal Name="OutputDataNameTemplate" OntologyEntryURI="terms.fhcrc.org#XarTemplate.OutputDataName" ValueType="String">MzXML file</exp:SimpleVal> </exp:ParameterDeclarations> </exp:Protocol> |
|
Example4.search.xar.xml
Two of the protocols in the generated xar.xml use the ${AutoFileLSID} template including the Convert to PepXml step shown. But note here that the OutputDataFileTemplate parameter is declared but does not have a default value. |
<exp:Protocol rdf:about="${FolderLSIDBase}:MS2.ConvertToPepXml"> <exp:Name>Convert To PepXml</exp:Name> <exp:ApplicationType>ProtocolApplication</exp:ApplicationType> <exp:MaxInputMaterialPerInstance>0</exp:MaxInputMaterialPerInstance> <exp:MaxInputDataPerInstance>1</exp:MaxInputDataPerInstance> <exp:OutputMaterialPerInstance>0</exp:OutputMaterialPerInstance> <exp:OutputDataPerInstance>1</exp:OutputDataPerInstance> <exp:ParameterDeclarations> <exp:SimpleVal Name="ApplicationLSIDTemplate" OntologyEntryURI="terms.fhcrc.org#XarTemplate.ApplicationLSID" ValueType="String">${RunLSIDBase}::MS2.ConvertToPepXml</exp:SimpleVal> <exp:SimpleVal Name="ApplicationNameTemplate" OntologyEntryURI="terms.fhcrc.org#XarTemplate.ApplicationName" ValueType="String">PepXml/XTandem Search Results</exp:SimpleVal> <exp:SimpleVal Name="OutputDataLSIDTemplate" OntologyEntryURI="terms.fhcrc.org#XarTemplate.OutputDataLSID" ValueType="String">${AutoFileLSID}</exp:SimpleVal> <exp:SimpleVal Name="OutputDataFileTemplate" OntologyEntryURI="terms.fhcrc.org#XarTemplate.OutputDataFile" ValueType="String"/> <exp:SimpleVal Name="OutputDataNameTemplate" OntologyEntryURI="terms.fhcrc.org#XarTemplate.OutputDataName" ValueType="String">PepXml/XTandem Search Results</exp:SimpleVal> </exp:ParameterDeclarations> <exp:Properties/> </exp:Protocol>
|
|
The StartingInputDefintions use the ${AutoFileLSID} template. This time the files referred to are in different directories from the xar.xml file. The Xar load process turns these relative paths into paths relative to the Pipeline root when checking to see if Data objects already point to them. |
<exp:StartingInputDefinitions> <exp:Data rdf:about="${AutoFileLSID}"> <exp:Name>Example4.mzXML</exp:Name> <exp:CpasType>Data</exp:CpasType> <exp:DataFileUrl>../../Example4.mzXML</exp:DataFileUrl> </exp:Data> <exp:Data rdf:about="${AutoFileLSID}"> <exp:Name>Tandem Settings</exp:Name> <exp:CpasType>Data</exp:CpasType> <exp:DataFileUrl>tandem.xml</exp:DataFileUrl> </exp:Data> <exp:Data rdf:about="${AutoFileLSID}"> <exp:Name>Bovine_mini.fasta</exp:Name> <exp:CpasType>Data</exp:CpasType> <exp:DataFileUrl>..\..\databases\Bovine_mini.fasta</exp:DataFileUrl> </exp:Data> </exp:StartingInputDefinitions>
|
|
The ExperimentLog section of this xar.xml uses the optional CommonParametersApplied element to give the values for the OutputDataFileTemplate parameters. This element has the effect of applying the same parameter values to all ProtocolApplications generated for the current action. |
<exp:ExperimentLog> <exp:ExperimentLogEntry ActionSequenceRef="1"/> <exp:ExperimentLogEntry ActionSequenceRef="30"> <exp:CommonParametersApplied> <exp:SimpleVal Name="OutputDataFileTemplate" OntologyEntryURI="terms.fhcrc.org#XarTemplate.OutputDataFile" ValueType="String">Example4.xtan.xml</exp:SimpleVal> </exp:CommonParametersApplied> </exp:ExperimentLogEntry> <exp:ExperimentLogEntry ActionSequenceRef="40"> <exp:CommonParametersApplied> <exp:SimpleVal Name="OutputDataFileTemplate" OntologyEntryURI="terms.fhcrc.org#XarTemplate.OutputDataFile" ValueType="String">Example4.pep.xml</exp:SimpleVal> </exp:CommonParametersApplied> </exp:ExperimentLogEntry> <exp:ExperimentLogEntry ActionSequenceRef="50"/> </exp:ExperimentLog> |
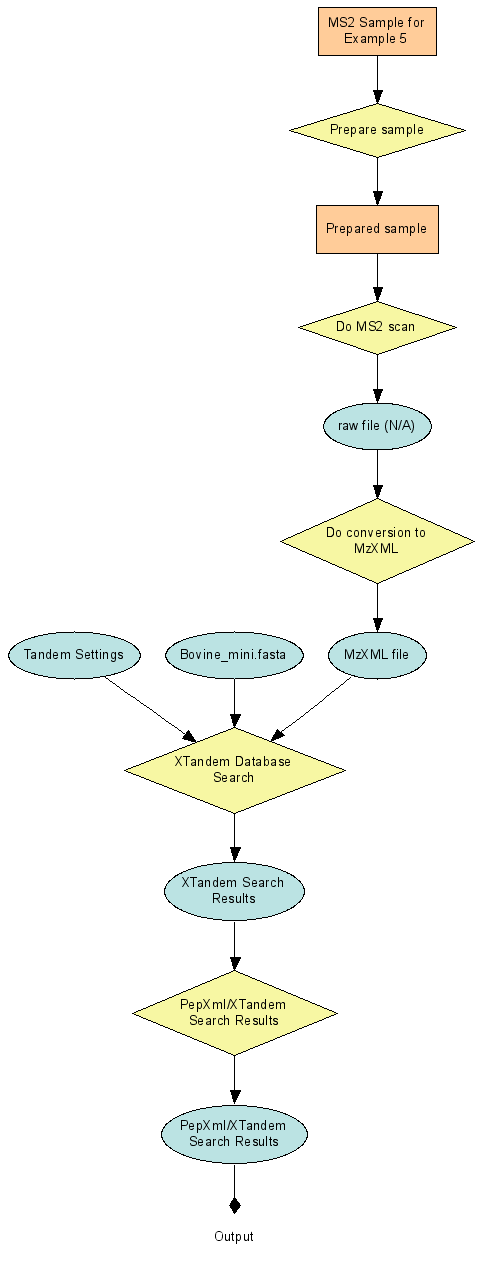
After using the Data Pipeline to generate a pep.xml peptide search result, some users may want to integrate the two separate connected runs of Example 4 into a single run that starts with a sample and ends with the peptide search results. Example 5 is the result of this combination.
Combine connected runs into an end-to-end run (Example 5)
|
|
|
|
Summary View |
Details View |
Table 5: Highlights of MS2 end-to-end experiment description (Example5.xar.xml)
|
The protocols of example 5 are the union of the two sets of protocols in Example4.xar.xml and Example4.search.xar.xml. A new run protocol becomes the parent of all of the steps.
Note that the ActionDefinition section has one unusual addition: the XTandemAnalyze step has both the MS2EndToEndProtocol (first) step and the ConvertToMzXML steps as predecessors. This is because it takes as inputs 3 files: the mzXML file output by step 30 and the tandem.xml and bovine_mini.fasta files. The latter two files are not produced by any step in the protocol and so must be included in the StartingInputs section. Adding step 1 as a predecessor is the signal that the XTandemAnalyze step uses StartingInputs. |
<exp:ProtocolActionDefinitions> <exp:ProtocolActionSet ParentProtocolLSID="${FolderLSIDBase}:MS2EndToEndProtocol"> <exp:ProtocolAction ChildProtocolLSID="${FolderLSIDBase}:MS2EndToEndProtocol" ActionSequence="1"> <exp:PredecessorAction ActionSequenceRef="1"/> </exp:ProtocolAction> <exp:ProtocolAction ChildProtocolLSID="${FolderLSIDBase}:SamplePrep" ActionSequence="10"> <exp:PredecessorAction ActionSequenceRef="1"/> </exp:ProtocolAction> <exp:ProtocolAction ChildProtocolLSID="${FolderLSIDBase}:LCMS2" ActionSequence="20"> <exp:PredecessorAction ActionSequenceRef="10"/> </exp:ProtocolAction> <exp:ProtocolAction ChildProtocolLSID="${FolderLSIDBase}:ConvertToMzXML" ActionSequence="30"> <exp:PredecessorAction ActionSequenceRef="20"/> </exp:ProtocolAction> <exp:ProtocolAction ChildProtocolLSID="${FolderLSIDBase}:XTandemAnalyze" ActionSequence="60"> <exp:PredecessorAction ActionSequenceRef="1"/> <exp:PredecessorAction ActionSequenceRef="30"/> </exp:ProtocolAction> <exp:ProtocolAction ChildProtocolLSID="${FolderLSIDBase}:ConvertToPepXml" ActionSequence="70"> <exp:PredecessorAction ActionSequenceRef="60"/> </exp:ProtocolAction> <exp:ProtocolAction ChildProtocolLSID="${FolderLSIDBase}:MarkRunOutput" ActionSequence="1000"> <exp:PredecessorAction ActionSequenceRef="70"/> </exp:ProtocolAction> </exp:ProtocolActionSet> </exp:ProtocolActionDefinitions> |
Describing pooling and fractionation
Some types of MS2 experiments involve combining two related samples into one prior to running LCMS2. The original samples are dyed with different markers so that they can be distinguished. Example 6 demonstrates how to do this in a xar.xml.
Sample pooling and fractionation (Example 6)
|
|
|
Details View |
Table 6: Describing pooling and fractionation (Example6.xar.xml)
|
There are two different tagging protocols for the two different dye types.
The PoolingTreatment protocol has a MaxInputMaterialPerInstance of 2 and an Output of 1
|
<exp:Protocol rdf:about="${FolderLSIDBase}:TaggingTreatment.Cy5"> <exp:Name>Label with Cy5</exp:Name> <exp:ProtocolDescription>Tag sample with Amersham CY5 dye</exp:ProtocolDescription> … </exp:Protocol> <exp:Protocol rdf:about="${FolderLSIDBase}:TaggingTreatment.Cy3"> <exp:Name>Label with Cy3</exp:Name> … </exp:Protocol> <exp:Protocol rdf:about="${FolderLSIDBase}:PoolingTreatment"> <exp:Name>Combine tagged samples</exp:Name> <exp:ProtocolDescription/> <exp:ApplicationType/> <exp:MaxInputMaterialPerInstance>2</exp:MaxInputMaterialPerInstance> <exp:MaxInputDataPerInstance>0</exp:MaxInputDataPerInstance> <exp:OutputMaterialPerInstance>1</exp:OutputMaterialPerInstance> <exp:OutputDataPerInstance>0</exp:OutputDataPerInstance> … </exp:Protocol> |
|
Both tagging steps are listed as having the start protocol (action sequence =1) as predecessors, meaning that they take StartingInputs.
The pooling step lists both the tagging steps as predecessors. |
<exp:ProtocolActionDefinitions> <exp:ProtocolActionSet ParentProtocolLSID="${FolderLSIDBase}:Example_6_Protocol"> <exp:ProtocolAction ChildProtocolLSID="${FolderLSIDBase}:Example_6_Protocol" ActionSequence="1"> <exp:PredecessorAction ActionSequenceRef="1"/> </exp:ProtocolAction> <exp:ProtocolAction ChildProtocolLSID="${FolderLSIDBase}:TaggingTreatment.Cy5" ActionSequence="10"> <exp:PredecessorAction ActionSequenceRef="1"/> </exp:ProtocolAction> <exp:ProtocolAction ChildProtocolLSID="${FolderLSIDBase}:TaggingTreatment.Cy3" ActionSequence="11"> <exp:PredecessorAction ActionSequenceRef="1"/> </exp:ProtocolAction> <exp:ProtocolAction ChildProtocolLSID="${FolderLSIDBase}:PoolingTreatment" ActionSequence="15"> <exp:PredecessorAction ActionSequenceRef="10"/> <exp:PredecessorAction ActionSequenceRef="11"/> </exp:ProtocolAction> |
|
The two starting inputs need to be assigned to specific steps so that the xar records which dye was applied to which sample. So this xar.xml uses the ApplicationInstanceCollection element of the ExperimentLogEntry to specify which input a step takes. Since there is only one instance of step 10 (or 20) there is one InstanceDetails block in the collection. The InstanceInputs refer to an LSID in the StartingInputDefinitions block. Instance-specific parameters could also be specified in this section. |
<exp:StartingInputDefinitions> <exp:Material rdf:about="${FolderLSIDBase}:Case"> <exp:Name>Case</exp:Name> </exp:Material> <exp:Material rdf:about="${FolderLSIDBase}:Control"> <exp:Name>Control</exp:Name> </exp:Material> </exp:StartingInputDefinitions>
<exp:ExperimentLog> <exp:ExperimentLogEntry ActionSequenceRef="1"/> <exp:ExperimentLogEntry ActionSequenceRef="10"> <exp:ApplicationInstanceCollection> <exp:InstanceDetails> <exp:InstanceInputs> <exp:MaterialLSID>${FolderLSIDBase}:Case</exp:MaterialLSID> </exp:InstanceInputs> </exp:InstanceDetails> </exp:ApplicationInstanceCollection> </exp:ExperimentLogEntry> <exp:ExperimentLogEntry ActionSequenceRef="11"> <exp:ApplicationInstanceCollection> <exp:InstanceDetails> <exp:InstanceInputs> <exp:MaterialLSID>${FolderLSIDBase}:Control</exp:MaterialLSID> </exp:InstanceInputs> </exp:InstanceDetails> </exp:ApplicationInstanceCollection> </exp:ExperimentLogEntry> <exp:ExperimentLogEntry ActionSequenceRef="15"/> |
Full Example: Lung Adenocarcinoma Study description
The file LungAdenocarcinoma.xar.xml is a fully annotated description of an actual study. It uses export format because it includes custom properties attached to run outputs. Properties of generated outputs cannot currently be described using log format.