Access Your Data as "labkey.data"
LabKey Server automatically reads your chosen dataset into a data frame called
labkey.data using
Input Substitution.
A data frame can be visualized as a list with unique row names and columns of consistent lengths. Column names are converted to all lower case, spaces or slashes are replaced with underscores, and some special characters are replaced with words (i.e. "CD4+" becomes "cd4_plus_"). You can see the column names for the built in labkey.data frame by calling:
options(echo=TRUE);
names(labkey.data);
Just like any other data.frame, data in a column of labkey.data can be referenced by the column's name, converted to all lowercase and preceded by a $:
labkey.data$<column name>
For example,
labkey.data$pulse; provides all the data in the Pulse column. Learn more about column references
below.
Note that the examples in this section frequently include column names. If you are using your own data or a different version of LabKey example data, you may need to retrieve column names and edit the code examples given.
Use Pre-existing R Scripts
To use a pre-existing R script with LabKey data, try the following procedure:
- Open the R Report Builder:
- Open the dataset of interest ("Physical Exam" for example).
- Select > Create R Report.
- Paste the script into the Source tab.
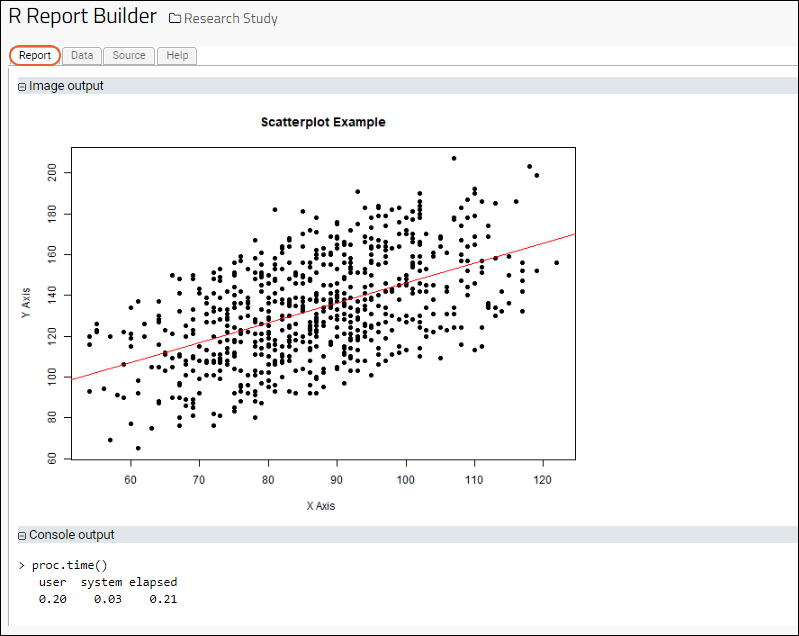
- Identify the LabKey data columns that you want to be represented by the script, and load those columns into vectors. The following loads the Systolic Blood Pressure and Diastolic Blood Pressure columns into the vectors x and y:
x <- labkey.data$diastolicbp;
y <- labkey.data$systolicbp;
png(filename="${imgout:myscatterplot}", width = 650, height = 480);
plot(x,
y,
main="Scatterplot Example",
xlab="X Axis ",
ylab="Y Axis",
pch=19);
abline(lm(y~x), col="red") # regression line (y~x);
- Click the Report tab to see the result:
Find Simple Means
Once you have loaded your data, you can perform statistical analyses using the functions/algorithms in R and its associated packages. For example, calculate the mean Pulse for all participants.
options(echo=TRUE);
names(labkey.data);
labkey.data$pulse;
a <- mean(labkey.data$pulse, na.rm= TRUE);
a;
Find Means for Each Participant
The following simple script finds the average values of a variety of physiological measurements for each study participant.
# Get means for each participant over multiple visits;
options(echo=TRUE);
participant_means <- aggregate(labkey.data, list(ParticipantID = labkey.data$participantid), mean, na.rm = TRUE);
participant_means;
We use na.rm as an argument to
aggregate in order to calculate means even when some values in a column are NA.
Create Functions in R
This script shows an example of how functions can be created and called in LabKey R scripts. Before you can run this script, the Cairo package must be installed on your server. See
Install and Set Up R for instructions.
Note that the second line of this script creates a "data" copy of the input file, but removes all participant records that contain an NA entry. NA entries are common in study datasets and can complicate display results.
library(Cairo);
data= na.omit(labkey.data);
chart <- function(data)
{
plot(data$pulse, data$pulse);
};
filter <- function(value)
{
sub <- subset(labkey.data, labkey.data$participantid == value);
#print("the number of rows for participant id: ")
#print(value)
#print("is : ")
#print(sub)
chart(sub)
}
names(labkey.data);
Cairo(file="${imgout:a}", type="png");
layout(matrix(c(1:4), 2, 2, byrow=TRUE));
strand1 <- labkey.data[,1];
for (i in strand1)
{
#print(i)
value <- i
filter(value)
};
dev.off();
Access Data in Another Dataset (Select Rows)
You can use the Rlabkey library's
selectRows to specify the data to load into an R data frame, including labkey.data, or a frame named something else you choose.
For example, if you use the following, you will load some example fictional data from our public demonstration site that will work with the above examples.
library(Rlabkey)
labkey.data <- labkey.selectRows(
baseUrl="https://www.labkey.org",
folderPath="/home/Demos/HIV Study Tutorial/",
schemaName="study",
queryName="PhysicalExam",
viewName="",
colNameOpt="rname"
)
Convert Column Names to Valid R Names
Include colNameOpt="rname" to have the selectRows call provide "R-friendly" column names. This converts column names to lower case and replaces spaces or slashes with underscores. Note that this may be different from the built in column name transformations in the built in labkey.data frame. The built in frame also substitutes words for some special characters, i.e. "CD4+" becomes "cd4_plus_", so during report development you'll want to check using
names(labkey.data); to be sure your report references the expected names.
Learn more in the
Rlabkey Documentation.
Select Specific Columns
Use the
colSelect option with to specify the set of columns you want to add to your dataframe. Make sure there are no spaces between the commas and column names.
In this example, we load some fictional example data, selecting only a few columns of interest.
library(Rlabkey)
labkey.data <- labkey.selectRows(
baseUrl="https://www.labkey.org",
folderPath="/home/Demos/HIV Study Tutorial/",
schemaName="study",
queryName="Demographics",
viewName="",
colSelect="ParticipantId,date,cohort,height,Language",
colFilter=NULL,
containerFilter=NULL,
colNameOpt="rname"
)
Display Lookup Target Columns
If you load the above example, and then execute:
labkey.data$language; you will see all the data in the "Language" column.
Remember that in an R data frame, columns are referenced in all lowercase, regardless of casing in LabKey Server. For consistency in your selectRows call, you can also define the colSelect list in all lowercase, but it is not required.
If "Language" were a lookup column referencing a list of Languages with accompanying translators, this would return a series of integers or whatever the key of the "Language" column is.
You could then access a column that is not the primary key in the lookup target, typically a more human-readable display values. Using this example, if the "Translator" list included "LanguageName, TranslatorName and TranslatorPhone" columns, you could use syntax like this in your selectRows,
library(Rlabkey)
labkey.data <- labkey.selectRows(
baseUrl="https://www.labkey.org",
folderPath="/home/Demos/HIV Study Tutorial/",
schemaName="study",
queryName="Demographics",
viewName="",
colSelect="ParticipantId,date,cohort,height,Language,Language/LanguageName,Language/TranslatorName,Language/TranslatorPhone",
colFilter=NULL,
containerFilter=NULL,
colNameOpt="rname"
)
You can now retrieve human-readable values from within the "Language" list by converting everything to lowercase and substituting an underscore for the slash. Executing
labkey.data$language_languagename; will return the list of language names.
Access URL Parameters and Data Filters
While you are developing your report, you can acquire any URL parameters as well as any filters applied on the
Data tab by using
labkey.url.params.
For example, if you filter the "systolicBP" column to values over 100, then use:
...your report will include:
$`Dataset.systolicBP~gt`
[1] "100"
Write Result File to File Repository
The following report, when run, creates a result file in the server's file repository. Note that fileSystemPath is an absolute file path. To get the absolute path, see
Using the Files Repository.
fileSystemPath = "/labkey/labkey/MyProject/Subfolder/@files/"
filePath = paste0(fileSystemPath, "test.tsv");
write.table(labkey.data, file = filePath, append = FALSE, sep = "t", qmethod = "double", col.names=NA);
print(paste0("Success: ", filePath));
Related Topics
