Label-Free Quantitation Using Spectra Counts
When given two unlabeled samples that are input to a mass spectrometer, it is often desirable to assess whether a given protein exists in higher abundance in one sample compared to the other. One strategy for doing so is to
count the spectra identified for each sample by the search engine. This technique requires a statistical comparison of multiple, repeated MS2 runs of each sample. LabKey Server makes handling the data from multiple runs straightforward.
Example Data Set
To illustrate the technique, we will use mzXML files that were described in
this paper as the "Variability Mix":
Jacob D. Jaffe, D. R. Mani, Kyriacos C. Leptos, George M. Church, Michael A. Gillette, and Steven A. Carr, "PEPPeR, a Platform for Experimental Proteomic Pattern Recognition", Molecular and Cellular Proteomics; 5: 1927 - 1941, October 2006The datasets are derived from two sample protein mixes, alpha and beta, with varied concentrations of a specific list of 12 proteins. The samples were run on a Thermo Fisher Scientific LTQ FT Ultra Hybrid mass spectrometer. The resulting datafiles were converted to the mzXML format that was downloaded from Tranche.
The files named VARMIX_A through VARMIX_E were replicates of the Alpha mix. The files named VARMIX_K through VARMIX_O were the Beta mix.
You can see the examples in our
online demo project.
Running the MS2 Search
The mzXML files provided with the PEPPeR paper included both MS1 and MS2 scan data. The first task is to get an MS2 search protocol that correctly identifies the 12 proteins spiked into the samples. The published data do not include the FASTA file to use as the basis of the search, so this has to be created from the descriptions in the paper. The paper did provide the search parameters used by the authors, but these were given for the SpectrumMill search engine, which is not freely available nor accessible from LabKey Server. So the SpectrumMill parameters are translated into their approximate equivalents on the X!Tandem search engine that is included with LabKey Server.
Creating the right FASTA file
The PEPPeR paper gives the following information about the protein database against which they conducted their search:
Data from the Scale Mixes and Variability Mixes were searched against a small protein database consisting of only those proteins that composed the mixtures and common contaminants… Data from the mitochondrial preparations were searched against the International Protein Index (IPI) mouse database version 3.01 and the small database mentioned above.The spiked proteins are identified in the paper by common names such as "Aprotinin". The paper did not give the specific protein database identifiers such as IPI numbers or SwissProt.. The following list of 13 SwissProt names is based on Expasy searches using the given common names as search terms. (Note that "alpha-Casein" became two SwissProt entries).
| Common Name | Organism | SprotName | Conc. In A | Conc. In B |
|---|
| Aprotinin | Cow | BPT1_BOVIN | 100 | 5 |
| Ribonuclease | Cow | RNAS1_BOVIN | 100 | 100 |
| Myoglogin | Horse | MYG_HORSE | 100 | 100 |
| beta-Lactoglobulin | Cow | LACB_BOVIN | 50 | 1 |
| alpha-Casein S2 | Cow | CASA2_BOVIN | 100 | 10 |
| alpha-Casein S1 | Cow | CASA1_BOVIN | 100 | 10 |
| Carbonic anhydrase | Cow | CAH2_BOVIN | 100 | 100 |
| Ovalbumin | Chicken | OVAL_CHICK | 5 | 10 |
| Fibrinogen beta chain | Cow | FIBB_BOVIN | 25 | 25 |
| Albumin | Cow | ALBU_BOVIN | 200 | 200 |
| Transferrin | Human | TRFE_HUMAN | 10 | 5 |
| Plasminogen | Human | PLMN_HUMAN | 2.5 | 25 |
| beta-Galactosidase | E. Coli | BGAL_ECOLI | 1 | 10 |
As in the PEPPeR study, the total search database consisted of
- The spiked proteins as listed in the table, using SwissProt identifiers
- The Mouse IPI fasta database, using IPI identifiers
- The cRAP list of common contaminants from www.thegpm.org, minus the proteins that overlapped with the spiked proteins (including other species versions of those spiked proteins. This list used a different format of SwissProt identifiers.
Using different identifier formats for the three sets of sequences in the search database had the side effect of making it very easy to distinguish expected from unexpected proteins.
Loading the PEPPeR data as a custom protein list
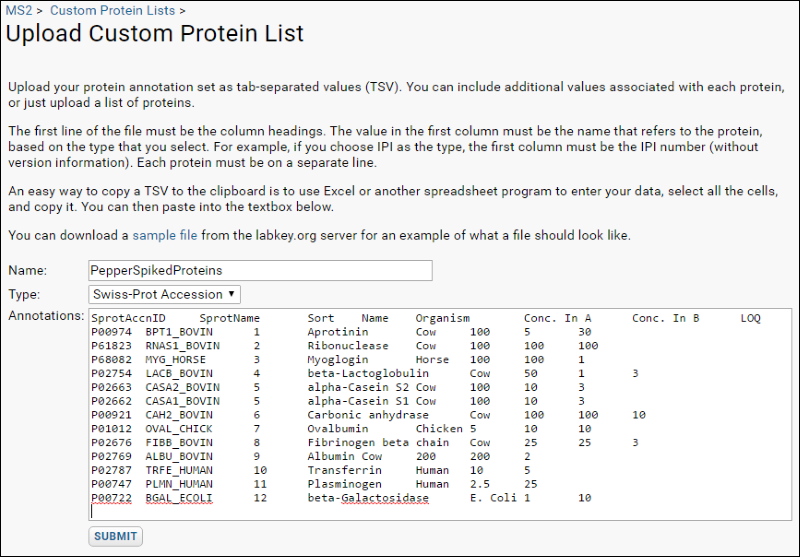
When analyzing a specific set of identified proteins as in this exercise, it is very useful to load the known data about the proteins as a custom protein annotation list. To add custom protein annotations using our example file attached to this page:
- Navigate to the MS2 Dashboard.
- Select Admin > Manage Custom Protein Lists.
- Click Import Custom Protein List.
- Download the attached file PepperProteins.tsv and open it.
- Select all rows and all columns of the content, and paste into the text box on the Upload Custom Protein Annotations page. The first column is a “Swiss-Prot Accession” value.
- Click Submit.
X!Tandem Search Parameters
Spectra counts rely on the output of the search engine, and therefore the search parameters will likely affect the results. The original paper used SpectrumMill and gave its search parameters. For LabKey Server, the parameters must be translated to X!Tandem. These are the parameters applied:
<bioml>
<!-- Carbamidomethylation (C) -->
<note label="residue, modification mass" type="input">57.02@C</note>
<!-- Carbamylated Lysine (K), Oxidized methionine (M) -->
<note label="residue, potential modification mass" type="input">43.01@K,16.00@M</note>
<note label="scoring, algorithm" type="input">k-score</note>
<note label="spectrum, use conditioning" type="input">no</note>
<note label="pipeline quantitation, metabolic search type" type="input">normal</note>
<note label="pipeline quantitation, algorithm" type="input">xpress</note>
</bioml>
Notes on these choices:
- The values for the fixed modifications for Carbamidomethylation and the variable modifications for Carbamylated Lysine (K) and Oxidized methionine (M) were taken from the Delta Mass database.
- Pyroglutamic acid (N-termQ) was another modification set in the SpectrumMill parameters listed in the paper, but X!Tandem checks for this modification by default.
- The k-score pluggable scoring algorithm and the associated “use conditioning=no” are recommended as the standard search configuration used at the Fred Hutchinson Cancer Research Center because of its familiarity and well-tested support by PeptideProphet.
- The metabolic search type was set to test the use of Xpress for label-free quantitation, but the results do not apply to spectra counts.
- These parameter values have not been reviewed for accuracy in translation from SpectrumMill.
Reviewing Search Results
One way to assess how well the X!Tandem search identified the known proteins in the mixtures is to compare the results across all 50 runs, or for the subsets of 25 runs that comprise the Alpha Mix set and the Beta Mix set. To enable easy grouping of the runs into Alpha and Beta mix sets, create two Run Groups (for example AlphaRunGroup and BetaRunGroup) and add the runs to them. Creating run groups is a sub function of the Add to run group button on the MS2 Runs (enhanced) grid.
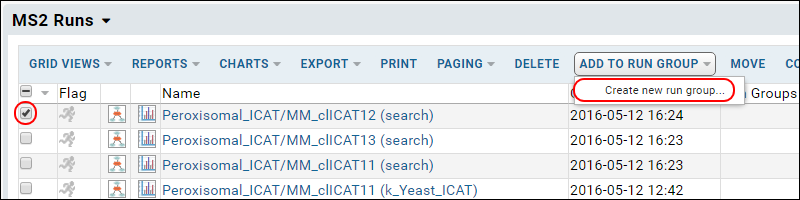
After the run groups have been created, and runs assigned to them, it is easy to compare the protein identifications in samples from just one of the two groups by the following steps:
- Navigate to the MS2 Dashboard and the MS2 Runs web part.
- If you do not see the Run Groups column, use Grid Views > Customize Grid to add it.
- Filter to show only the runs from one group by clicking it's name in the Run Groups column. If the name is not a link, you can use the column header filter option as usual.
- Select all the filtered runs using the checkbox at the top of the selection box column.
- Select Compare > ProteinProphet.
- On the options page choose "Peptides with PeptideProphet probability >=" and enter ".75".
- Click Compare.
The resulting comparison view will look something like this. You can customize this grid to show other columns as desired.
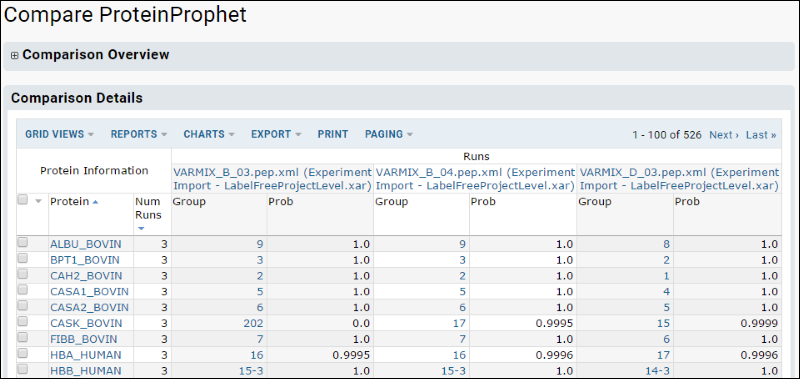
Most of the spiked proteins will show up in all 50 runs with a probability approaching 1.0. Two of the proteins, eta-Galactosidase and Plasminogen, appear in only half of the A mix runs. This is consistent with the low concentration of these two proteins in the Alpha mix as shown in t he table in an earlier section. Similarly, only beta-Lactoglobulin and Aprotinin fail to show up in all 25 of the runs for the B mix. These two are the proteins with the lowest concentration in beta.
Overall, the identifications seem to be strong enough to support a quantitation analysis.
The Spectra Count views
The wide format of the ProteinProphet view is designed for viewing on-line. It can be downloaded to an Excel or TSV file, but the format is not well suited for further client-side analysis after downloading. For example, the existence of multiple columns of data under each run in Excel makes it difficult to reference the correct columns in formulas. The spectra count views address this problem. These views have a regular column structure with Run Id as just a single column.
- Return to the MS2 Runs web part on the MS2 Dashboard and select the same filtered set of runs.
- Select Compare > Spectra Count.
The first choice to make when using the spectra count views is to decide what level of grouping to do in the database prior to exporting the dataset. The options are:
- Peptide sequence: Results are grouped by run and peptide. Use this for quantitation of peptides only
- Peptide sequence, peptide charge: Results grouped by run and peptide charge. Used for peptide quantitation if you need to know the charge state (for example, to filter or weight counts based on charge.state)
- Peptide sequence, ProteinProphet protein assignment: The run/peptide grouping joined with the ProteinProphet assignment of proteins for each peptide.
- Peptide sequence, search engine protein assignment: The run/peptide grouping joined with the single protein assigned by the search engine for each peptide.
- Peptide sequence, peptide charge, ProteinProphet protein assignment: Adds in grouping by charge state
- Peptide sequence, peptide charge, search engine protein assignment: Adds in grouping by charge state
- Search engine protein assignment: Grouped by run/protein assigned by the search engine.
- ProteinProphet protein assignment: Grouped by run/protein assigned by ProteinProphet. Use with protein group measurements generated by ProteinProphet
After choosing the grouping option, you also have the opportunity to filter the peptide-level data prior to grouping (much like a WHERE clause in SQL operates before the GROUP BY).
After the options page, LabKey Server displays the resulting data grouped as specified. Selecting
Grid Views > Customize Grid gives access to the column picker for choosing which data to aggregate, and what aggregate function to use. You can also specify a filter and ordering; these act after the grouping operation in the same way as SQL HAVING and ORDER BY apply after the GROUP BY.
Understanding the spectra count data sets
Because the spectra count output is a single rectangular result set, there will be repeated information with some grouping options. In the peptide, protein grid, for example, the peptide data values will be repeated for every protein that the peptide could be matched to. The table below illustrates this type of grouping:
| (row) | Run Id | Alpha Run Grp | Peptide | Charge States Obsv | Tot Peptide Cnt | Max PepProph | Protein | Prot Best Gene Name |
|---|
| 1 | 276 | false | K.AEFVEVTK.L | 2 | 16 | 0.9925 | ALBU_BOVIN | ALB |
| 2 | 276 | false | K.ATEEQLK.T | 2 | 29 | 0.9118 | ALBU_BOVIN | ALB |
| 3 | 276 | false | K.C^CTESLVNR.R | 1 | 18 | 0.9986 | ALBU_BOVIN | ALB |
| 4 | 276 | false | R.GGLEPINFQTAADQAR.E | 1 | 4 | 0.9995 | OVAL_CHICK | SERPINB14 |
| 5 | 276 | false | R.LLLPGELAK.H | 1 | 7 | 0.9761 | H2B1A_MOUSE | Hist1h2ba |
| 6 | 276 | false | R.LLLPGELAK.H | 1 | 7 | 0.9761 | H2B1B_MOUSE | Hist1h2bb |
| 7 | 276 | false | R.LLLPGELAK.H | 1 | 7 | 0.9761 | H2B1C_MOUSE | Hist1h2bg |
| 8 | 299 | true | K.AEFVEVTK.L | 2 | 16 | 0.9925 | ALBU_BOVIN | ALB |
| 9 | 299 | true | K.ECCHGDLLECADDR.A | 1 | 12 | 0.9923 | ALBU_MOUSE | Alb |
| 10 | 299 | true | R.LPSEFDLSAFLR.A | 1 | 1 | 0.9974 | BGAL_ECOLI | lacZ |
| 11 | 299 | true | K.YLEFISDAIIHVLHSK.H | 2 | 40 | 0.9999 | MYG_HORSE | MB |
In this example,
- Row 1 contains the total of all scans (16) that matched the peptide K.AEFVEVTK.L in Run 276, which was part of the Beta Mix. There were two charge states identified that contributed to this total, but the individual charge states are not reported separately in this grouping option. 0.9925 was the maximum probability calculated by PeptideProphet for any of the scans matched to this peptide The K.AEFVEVTK.L is identified with the ALBU_BOVIN (bovine albumin), which has a gene name of ALB.
- Rows 2 and 3 are different peptides in run 276 that also belong to Albumin.. Row 4 matches to a different protein, ovalbumin.
- Rows 5-7 are 3 different peptides in the same run that could represent any one of 3 mouse proteins, H2B1x_MOUSE. ProteinProphet assigned all three proteins into the same group. Note that the total peptide count for the peptide is repeated for each protein that it matches. This means that simply adding up the total peptide counts would over count in these cases. This is just the effect of a many-to-many relationship between proteins and peptides that is represented in a single result set.
- Rows 8-11 are from a different run that was done from an Alpha mix sample.
Using Excel Pivot Tables for Spectra Counts
An Excel pivot table is a useful tool for consuming the datasets returned by the Spectra count comparison in LabKey Server. It is very fast, for example, for rolling up the Protein grouping data set and reporting ProteinProphet’s “Total Peptides” count, which is a count of spectra with some correction for the potential pitfalls in mapping peptides to proteins.
Using R scripts for spectra counts
The spectra count data set can also be passed into an R script for statistical analysis, reporting and charting. R script files which illustrate this technique can be downloaded here. Note that column names are hard coded and may need adjustment to match your data.
 After the run groups have been created, and runs assigned to them, it is easy to compare the protein identifications in samples from just one of the two groups by the following steps:
After the run groups have been created, and runs assigned to them, it is easy to compare the protein identifications in samples from just one of the two groups by the following steps: