[Tutorial] [
General MS2 Demo] [
Label Free Quantitation Demo]
[
Team]
LabKey Server's Proteomics tools (formerly the Computational Proteomics Analysis System, "CPAS") provide a web-based system for managing, analyzing, and sharing high volumes of tandem mass spectrometry data. It employs open-source tools provided by the Trans Proteomic Pipeline, developed by the
Institute for Systems Biology.
Searches are performed against FASTA sequence databases using a peptide search tool such as X! Tandem, Sequest, Mascot, or Comet. Once the spectra have been searched and scored, results are typically analyzed by validation tools PeptideProphet and ProteinProphet. You can also configure LabKey to perform quantitation analyses on the scored results, using XPRESS or Q3.
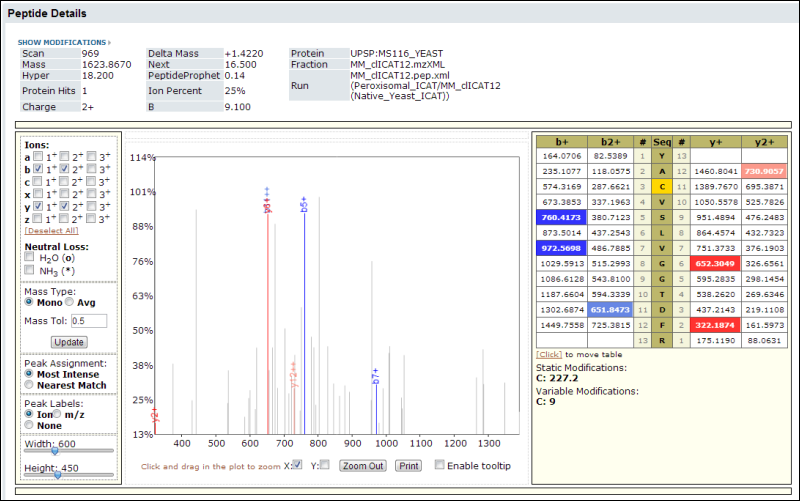
Analyzed results can be dynamically displayed, enabling you to filter, sort, customize, compare, and export experiment runs. You can share data securely with collaborators inside or outside your organization, with fine-grained control over permissions.
A
data pipeline imports and processes MS/MS data from raw and mzXML data files. The pipeline searches the data file for peptides using the X!Tandem search engine against the specified FASTA database. Once the data has been searched and scored (using X! Tandem scoring or a pluggable scoring algorithm), the pipeline optionally runs PeptideProphet, ProteinProphet, and XPRESS quantitation analyses on the search results.
The data pipeline can also load results that have been processed externally by some other programs. For example, it can load quantitation data processed by Q3.
Documentation Topics
Proteomics Installations
LabKey Server powers proteomics repositories at the following institutions:
Integrated Tools

 Analyzed results can be dynamically displayed, enabling you to filter, sort, customize, compare, and export experiment runs. You can share data securely with collaborators inside or outside your organization, with fine-grained control over permissions.A data pipeline imports and processes MS/MS data from raw and mzXML data files. The pipeline searches the data file for peptides using the X!Tandem search engine against the specified FASTA database. Once the data has been searched and scored (using X! Tandem scoring or a pluggable scoring algorithm), the pipeline optionally runs PeptideProphet, ProteinProphet, and XPRESS quantitation analyses on the search results.The data pipeline can also load results that have been processed externally by some other programs. For example, it can load quantitation data processed by Q3.
Analyzed results can be dynamically displayed, enabling you to filter, sort, customize, compare, and export experiment runs. You can share data securely with collaborators inside or outside your organization, with fine-grained control over permissions.A data pipeline imports and processes MS/MS data from raw and mzXML data files. The pipeline searches the data file for peptides using the X!Tandem search engine against the specified FASTA database. Once the data has been searched and scored (using X! Tandem scoring or a pluggable scoring algorithm), the pipeline optionally runs PeptideProphet, ProteinProphet, and XPRESS quantitation analyses on the search results.The data pipeline can also load results that have been processed externally by some other programs. For example, it can load quantitation data processed by Q3.