Luminex® analysis in LabKey Server makes use of a transform script to do a number of calculations including curve fits and estimated concentrations. To get the transform script running, you will need to:
Install and Configure R
You will need to install R and configure it as a scripting language on your LabKey Server. If you are on a Windows machine, install R in a directory that does not contain a space (i.e. not the default "C:\Program Files\R" location).
Install Necessary R Packages
The instructions in this section describe package installation using the R graphical user interface.
- Open your R console.
- Using the R console, install the packages listed below using commands like the following (you may want to vary the value of repos depending on your geographic location):
install.packages("Rlabkey", repos="http://cran.rstudio.com")
- Install the following packages (some may be downloaded for you as dependencies of others):
- Rlabkey
- xtable
- drc (will download additional packages you need)
- As an alternative to the R console, you can use the R graphical user interface:
- Use the drop-down menus to select Packages > Install package(s)...
- Select your CRAN mirror.
- Select the packages listed above. (You may be able to multi-select by using the Ctrl key.)
- Click OK and confirm that all packages were successfully unpacked and checked.
Obtain the Example Luminex Transform Script
The transform script example that LabKey uses in testing can be downloaded here. Also download the utility script "youtil.R":
Control Access to Scripts
Next, place the transform script and utility script in a server-accessible, protected location. For example, if your server runs on Windows you could follow these steps:
- Locate the <LK_ENLISTMENT> directory on your local machine. For example, it might be C:\labkey\labkeyEnlistment
- Create a new directory named scripts here.
- Place a copy of each of the files you downloaded in this new directory:
- labkey_luminex_transform.R
- youtil.R
For more information about locating R scripts, see
Transformation Scripts in R.
Associate the Transform Script with the Assay Design
Add Path to Assay Design
Edit the transform script path in the assay design we provided to point to this location.
- In the Assay List of your Luminex folder, click Luminex Assay 200.
- Click Manage Assay Design > Edit assay design.
- In the Assay Properties section, for the Transform Script, enter the full path to the scripts you just placed.
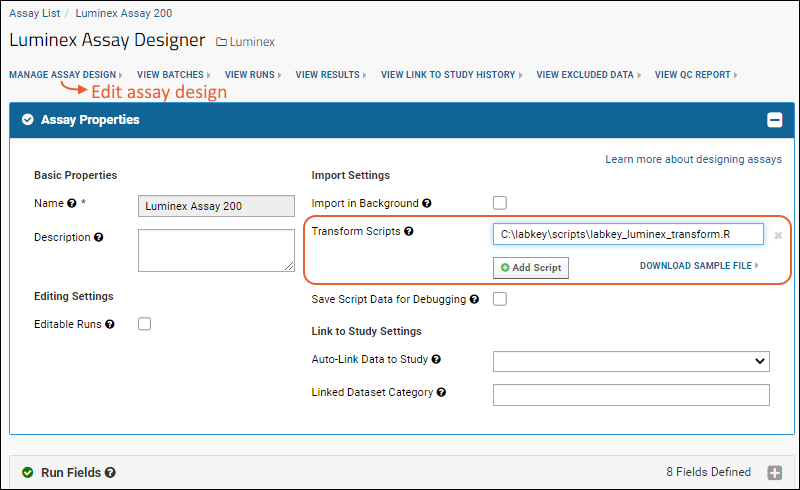
When you save, the server will verify the script location.
Test Package Configuration
This step is optional, but will confirm that your R script will run as needed in the following tutorial steps.
- Click Luminex.
- In the Files web part, select /Luminex/Runs - Assay 200/02-14A22-IgA-Biotin.xls.
- Click Import Data.
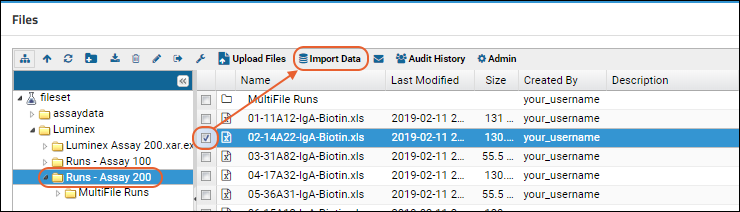
- Select Use Luminex Assay 200 in the popup and click Import again.
- For Batch properties, leave defaults unchanged and click Next.
- For Run Properties, leave defaults unchanged and click Next.
- Click Save and Finish.
If there is a problem with the path, or with installed packages or the version of R, error messages will help you figure out what else you need to do (e.g., installing an additional R package or upgrading your version of R). After installing a missing package, you can refresh your browser window to see if additional errors are generated.
If your script cannot find youtil.R, make sure it is located in the same directory as the LabKey Luminex transform script. The following should be peers:
- labkey_luminex_transform.R
- youtil.R
For further troubleshooting tips, see:
Troubleshoot Luminex Transform Scripts and Curve Fit ResultsWhen you see the Luminex Assay 200 Runs grid, you know your script was successfully run while importing the test run.
Delete the Imported Run Data
Before continuing with the tutorial, you need to delete the run you used to test your R configuration.
- Select > Manage Assays.
- Select Luminex Assay 200.
- Click the top checkbox on the left of the list of runs. This selects all rows.
- Click (Delete).
- Click Confirm Delete.

