LabKey Server allows you to quickly search for specific peptide identifications within the search results that have been loaded into a folder.
Performing a Peptide Search
There are a number of different places where you can initiate a search. If your folder is configured as an MS1 or MS2 folder, there may be a Peptide Search web part on the MS1 or MS2 Dashboard. You can also add the
Peptide Search web part to the portal page yourself. In some configurations, there may be a
Manage Peptide Inventory web part configured to allow searching and pooling of peptides.
Type in the peptide sequence to find. You may include modification characters if you wish. If you select the Exact Match checkbox, your results will only include peptides that match the exact peptide sequence, including modification characters.
Understanding the Search Results
The results page is divided into two sections.
The top section shows all of the MS1 features that have been identified, linked to MS2 peptides that match the search sequence, and loaded.
The bottom section shows all of the MS2 peptide identifications that match the search criteria, regardless of whether they match MS1 features.
You can apply filters to either section, customize the view to add or remove columns, or export them for analysis on other tools.
Mass Spec Search Web Part
If you will be searching for both proteins and peptides in a given folder, you may find it convenient to add the
Mass Spec Search (Tabbed) web part which combines
Protein Search and
Peptide Search in a single tabbed web part.
MS2 Runs With Peptide Counts
The MS2Extensions module contains an additional
MS2 Runs With Peptide Counts web part offering enhanced protein search capabilities, including filtering by multiple proteins simultaneously and the ability to focus on high-scoring identifications by using peptide filters.
The ms2extensions module is open source, but not shipped with the standard distribution.
Contact us to obtain it.
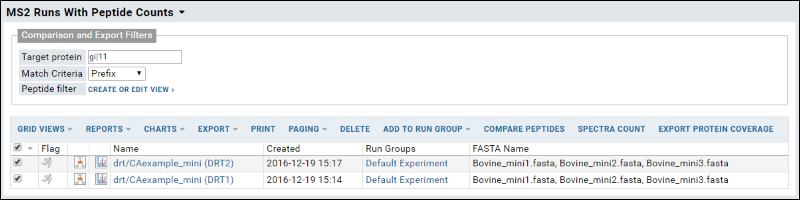
The runs list is preceded by a section for defining
Comparison and Export Filters:
- Target Protein: Enter one or more comma separated strings to identify proteins of interest.
- Match Criteria: Select how to match the above string(s). Options: Exact, Prefix, Suffix, Substring.
- Peptide Filter: Click Create or Edit View to define a peptide filter.
Select runs of interest with desired comparison filters and click any of the actions (
Compare Peptides, Spectra Count, or Export Protein Coverage. When multiple proteins match your search, you will see a
Disambiguate Proteins page. Use checkboxes to select applicable proteins. Hover over the triangle to see a detail panel about any protein to assist in selection.
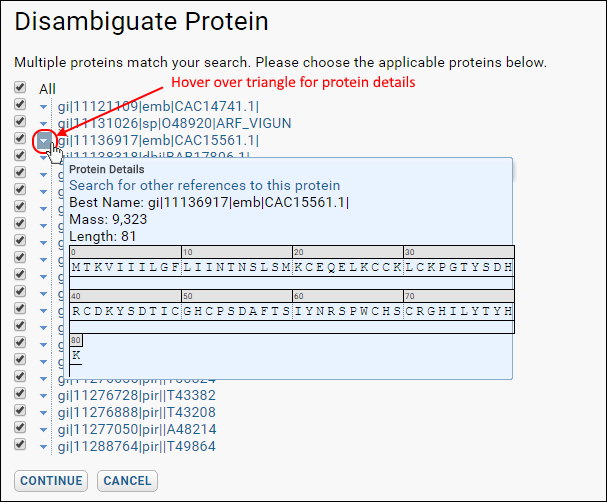
Click
Continue.
 Click Continue.
Click Continue.